The vascular disorder Hereditary Haemorrhagic Telangiectasia, more commonly known as Rendu-Osler disease, causes nosebleeds and arteriovenous malformations of the lungs, liver and central nervous system. This rare genetic disease is caused by mutations in several genes, including the Activin receptor-Like Kinase 1 (ALK1) gene, which codes for a receptor kinase expressed on the surface of the endothelial cells lining the blood vessels. Our team is seeking to gain a better understanding of the molecular mechanisms behind Rendu-Osler disease by studying the signalling pathways within the endothelial cells.
In 2007, our team identified two Bone Morphogenetic Proteins (BMP9 and BMP10) as ligands for the ALK1 receptor [1,2]. The binding of these two proteins to this receptor leads to the
phosphorylation of DNA transcription factors, whose function is to regulate gene expression. The aim of this work was to identify new signalling pathways activated by these factors by stimulating human endothelial cells. For the first time, a large-scale study in
collaboration made it possible to quantitatively compare the relative abundances of more than 10,000 phosphorylation sites. This phospho-proteomic analysis, based on
mass spectrometry, coupled with bioinformatic analyses made it possible to identify new signalling pathways in response to stimulation by the BMP9 and BMP10 proteins. In particular, the MAPK (mitogen activated protein kinases) signalling pathway [3].
The aim of this research is to propose new therapeutic ways for Rendu-Osler disease.
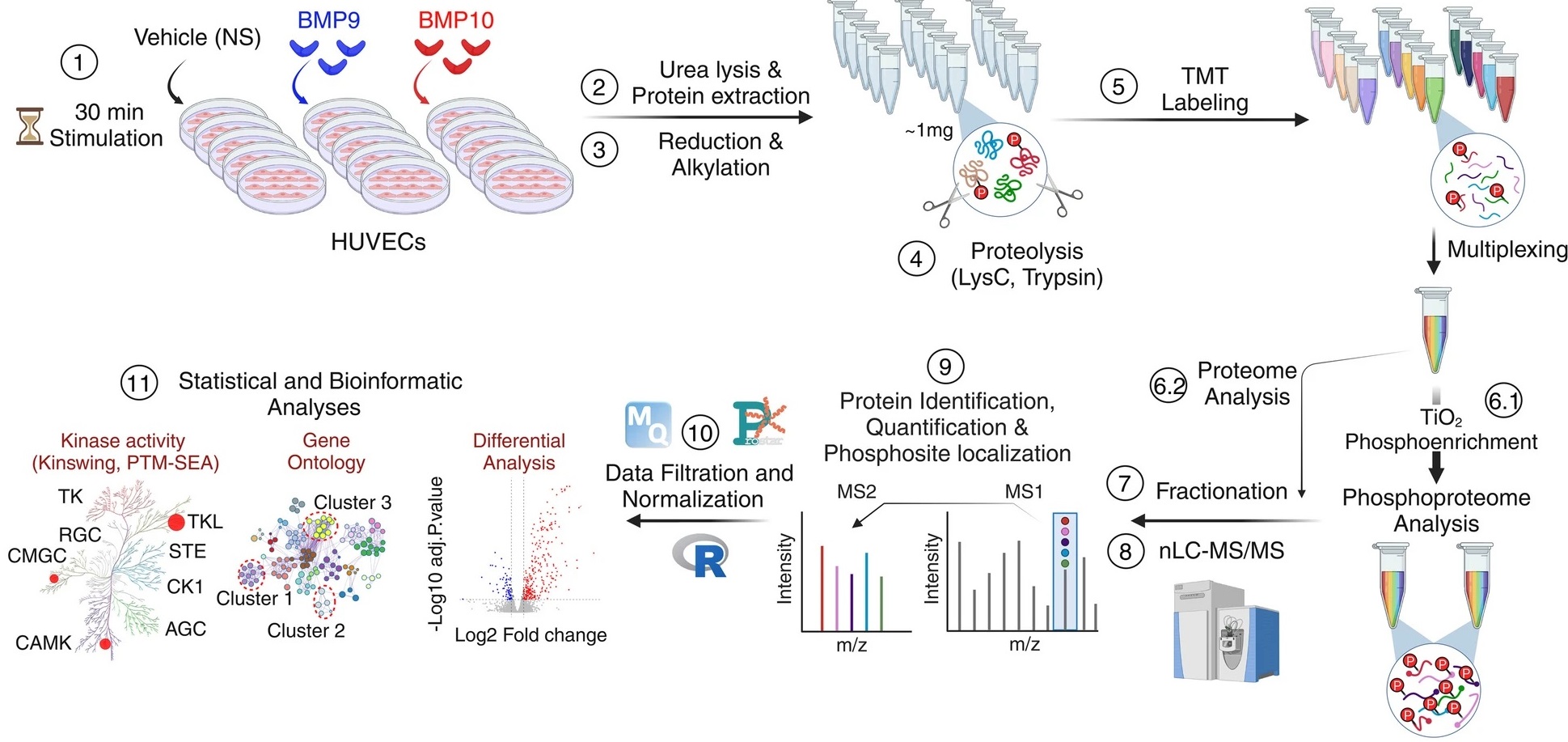
Figure: Endothelial cells are stimulated with BMP9 or BMP10 proteins and then treated to be analyzed using a phospho-proteomic approach based on mass spectrometry followed by bioinformatics analyses in order to identify differentially phosphorylated proteins and characterize new signaling pathways.
(1) Human umbilical cord endothelial cells (HUVEC) were stimulated or not (NS) with 10 ng/mL of BMP9 or BMP10 for 30 min.
(2) Lysates from five biological replicates per condition were prepared and
(3) subjected to reduction and alkylation, followed by
(4) digestion using a combination of two endoproteinases, LysC and trypsin. (5) The resulting peptides were labelled with tandem mass tag (TMT) reagents and pooled for analysis.
(6.1) The phosphorylated peptides were then enriched using titanium dioxide (TiO2) beads.
(6.2) while a small proportion of the pooled samples were reserved for proteomic analysis.
(7-8) Proteome and phospho-proteome samples were fractionated and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
(9–11) The data was then analyzed using a range of bioinformatics tools. Credit CEA
Phosphorylation is the addition of a phosphate group to a protein or small molecule, for example by a protein kinase.
Mass spectrometry is an analytical technique for determining molecular weight and identifying molecular complexes.
Collaboration
BAL (Biosanté) EDyP (Irig/BGE) and IMAC (Irig/Biosanté)